Novel Method for Accurately Predicting Cell Abundance Across Multiple Tumor Types
Date:15-11-2024 | 【Print】 【close】
Recently, a research team led by LI Xuefei at the Shenzhen Institutes of Advanced Technology (SIAT) of the Chinese Academy of Sciences, in collaboration with TIAN Liang’s team from the Hong Kong Baptist University, developed a deconvolution algorithm called DeSide. This algorithm is based on deep learning and publicly available scRNA-seq datasets, which can accurately estimate the abundance of 16 cell types across 19 types of solid tumors.
The sduty was published in PNAS on Nov. 8.
Solid tumors consist of various cell types, including cancer cells, endothelial cells, immune cells, and fibroblasts, etc. Previous studies show that the proportions of these cell types can strongly correlate with cancer progression, highlighting the importance of accurately assessing tumor cellular composition for clinical insights and therapy.
Current methods for investigating tumor cellular composition, such as flow cytometry and single-cell RNA sequencing (scRNA-seq), are typically costly and may suffer from low extraction efficiency. Consequently, series of studies have aimed to estimate cellular compositions in bulk RNA-seq samples. However, existing methods often perform inconsistently across different tumor types.
To address the challenge, the authors integrated 12 single-cell RNA-seq datasets from 6 solid tumor types, creating a comprehensive reference for synthesizing virtual bulk RNA-seq data (training set). They then introduced an innovative sampling method to generate virtual bulk tumors with a wider range of cell proportion combinations.To enhance the quality of synthesized data, they filtered genes and synthesized gene expression profiles, reducing input data complexity and increasing similarity to real tumor expression patterns.
Furthermore, DeSide introduces a novel architecture of the deep neural network, utilizing two fully connected networks to extract information from both biological signaling pathways and gene expression profiles.Moreover, DeSide refines the network’s activation function, effectively reducing prediction errors for tumor cell proportions.
As a result, DeSide outperforms existing methods in predicting cell abundance across 17 tumor types, showing significantly better performance in specific tumors compared to current referenced-base models. Additionally, DeSide can accurately predicts cellular compositions for cancer types absent from the training set, demonstrating strong generalization potential.
This study offers robust methods to better understand the tumor microenvironment, enhance prognostic evaluation, and facilitate the development of targeted therapies.
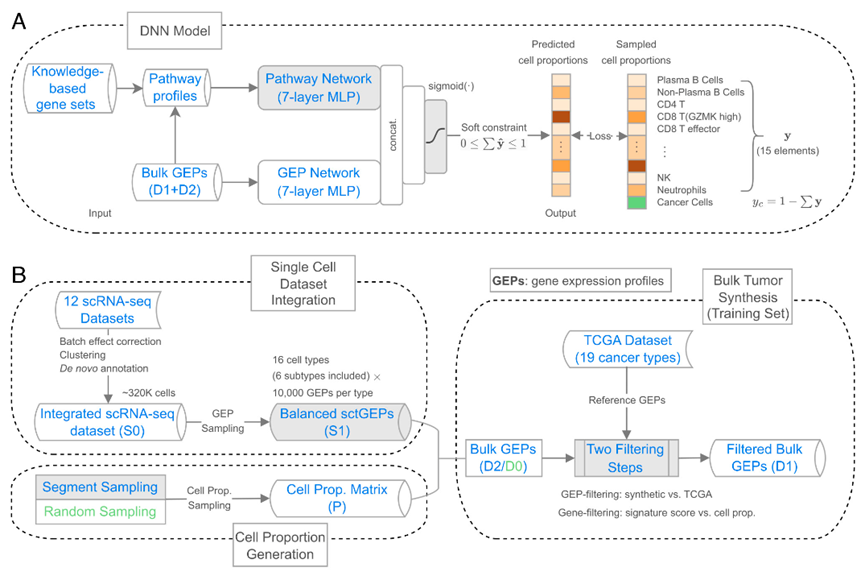
Overview of DeSide. (A) The deep neural network (DNN) model; (B) Bulk tumor synthesis workflow. (Image by SIAT)
Media Contact: LU Qun
Email: qun.lu@siat.ac.cn
Download the attachment:
DeSide: A unified deep learning approach for cellular deconvolution of tumor microenvironment