Scientists Develop Efficient Method for Reshaping Yeast DNA
Date:11-03-2024 | 【Print】 【close】
Chinese scientists, along with their international counterparts from the University of Manchester, have developed a method called construction of a repetitive array by transformation (CAT) for efficiently inserting rDNA arrays at any designated sites, which reshaped the ribosomal rDNA organization in budding yeast.
The research team from the Shenzhen Institute of Advanced Technology (SIAT) of Chinese Academy of Sciences, recently published their research in Cell Reports on Feb. 6.
In eukaryotes, the arrangement of rDNA genes commonly consists of clusters composed of tandem repeats. The repetitive nature and high transcription rates make rDNA one of the most unstable genomic regions, posing a significant challenge for study.
"Redundancy is a common feature of genomes," said Dr. JIANG Shuangying, the first author of this study, "One challenging aim of synthetic genomics is to devise a minimal genome that contains only the genes required to sustain free-living self-replication, aiming to help understand the core functions of life."
In this study, the entire native rDNA array on chromosome XII (ChrXII) in budding yeast was eliminated, and a synthetic rDNA array containing a loxPsym site within each rDNA unit was constructed on ChrIII. Subsequent to the introduction of Cre recombinase into the cells, the rDNA copy number was reduced to only 8 copies, lower than the previously reported copy number required for cell viability.
Furthermore, researchers constructed a series of strains containing two or three rDNA arrays using CAT to mimic rDNA organization in higher organisms. Results showed that the strains containing additional synthetic rDNA arrays exhibited no growth deficiencies under nutrient-rich conditions and all displayed a crescent-shaped nucleolus.
Despite dramatically altering the three-dimensional genome structure, these additional rDNA arrays had a minimal impact on global gene transcription.
This work sheds light on the high plasticity of rDNA organization and paves the way for the future rDNA engineering.
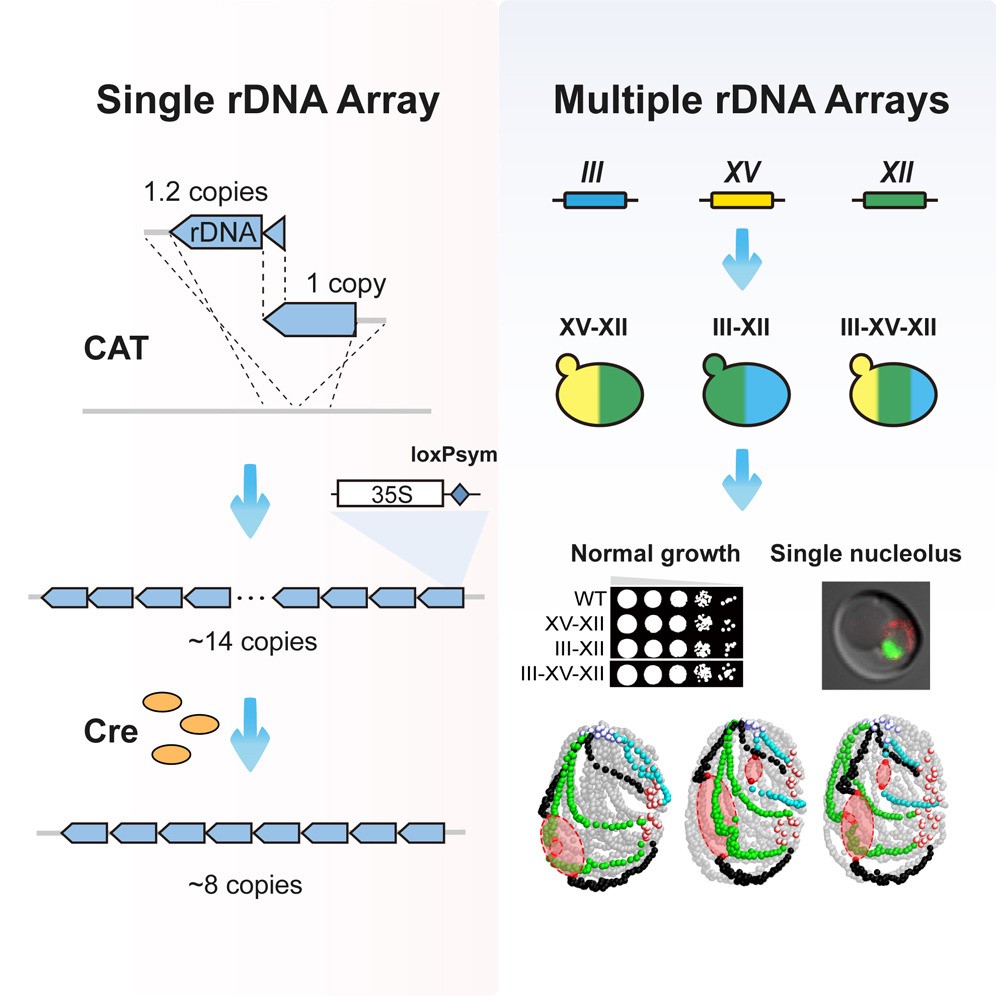
Fig. 1 Schematic Overview: Construction and reduction of a synthetic rDNA array (left); Construction and analysis of yeast strains containing multiple rDNA arrays (right). (Image by SIAT)
Fig.2 Cover: Introduction of an artificial control system for rDNA copy number. (Image by SIAT)
Media Contact:
ZHANG Xiaomin
Email:xm.zhang@siat.ac.cn