Researchers Develop Dynamic Model Integrating Protein Allocation with Flux Balance Analysis
Date:05-03-2024 | 【Print】 【close】
Recently, a research team led by Prof. FU Xiongfei from the Shenzhen Institute of Advanced Technology of Chinese Academy of Sciences has developed a dynamic model integrating protein allocation with flux balance analysis, which can predict the metabolic flux distribution during the nutrient shifts even without the detailed information of enzyme kinetics.
The study was published in Metabolic Engineering.
The natural habitats of microbes are characterized by constant exposure to dynamic conditions. In order to effectively adapt to fluctuating environments, such as shifts in nutrient availability, microbes typically undergo simultaneous redistribution of their metabolic fluxes and reallocation of protein synthesis.
However, predicting the coordination of cellular adaptations remains a significant challenge in systems biology due to the intricate cross-regulation between cellular metabolic fluxes and protein synthesis.
In this study, researchers developed a dynamic Constrained Allocation Flux Balance Analysis (dCAFBA) method, which integrates flux-controlled protein synthesis and enzyme constrained flux balance analysis. This approach was used to predict the transition kinetics of the metabolic fluxes of E.coli and discover the bottleneck metabolic reactions during the growth adaption.
Comparing the metabolic fluxes predicted by dCAFBA with the published proteome data, researchers found that during nutrient down-shifts, a switch of metabolic bottleneck from carbon uptake proteins to metabolic enzymes, which disrupts the coordination between metabolic fluxes and their enzyme abundance. This bottleneck switch results in a growth overshoot which was overlooked in previous studies.
Furthermore, the researchers extended dCAFBA to explore the cellular dynamics perturbed by the inducible exogenous gene expression for lycopene production in E.coli. Results showed that increasing levels of gene induction do indeed lead to higher amounts of product accumulation during a specific time frame, but the incremental gains diminished as induction intensity rises. These findings offer theoretical guidance for designing genetic circuits to enable real-time control of key enzyme expression and to maximize metabolite yields.
This work provides a quantitative tool to investigate cellular metabolism under varying environments and reveals insights into bacterial adaptation strategies.
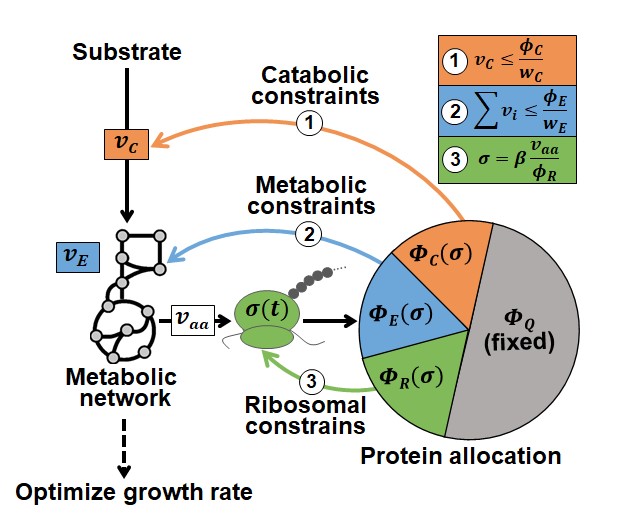
Illustration of the dCAFBA method. (Image by SIAT)
Media Contact:
ZHANG Xiaomin
Email:xm.zhang@siat.ac.cn