New Genome Editing Tools for Human Gut Microbiome
Date:13-01-2022 | 【Print】 【close】
Bacteroides is the most abundant genus in the human gut microbiome and is regarded as a “window” into the gut microbiome. Bacteroides species have been associated with various diseases, such as obesity, inflammatory bowel disease, colorectal cancer and so on.
Genome editing tools are essential for studying the function of gut commensal microbes within host health. However, existing genetic tools for human gut Bacteroides are inconvenient and have low editing efficiency.
On January 6th, the research group led by Prof. DAI Lei at the Shenzhen Institute of Advanced Technology (SIAT) of the Chinese Academy of Sciences, published their work “CRISPR/Cas-based genome editing for human gut commensal Bacteroides species" in ACS Synthetic Biology. ZHENG Linggang and TAN Yang are co-first authors.
In this paper, the researchers presented a versatile and efficient CRISPR/Cas-based editing tool for human gut Bacteroides (Figure 1). This genome editing tool is streamlined, markerless, highly efficient and easily followed.
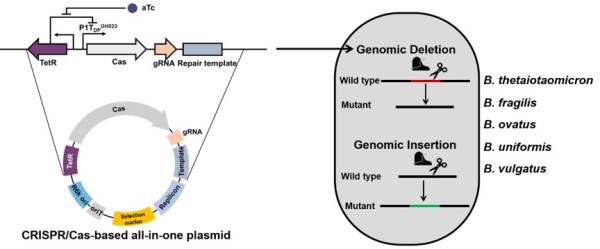
Fig. 1 CRISPR/Cas-based genome editing tool for human gut Bacteroides (Image by SIAT)
The researchers constructed multiple CRISPR/Cas systems in all-in-one Bacteroides-E. coli shuttle plasmids and systematically evaluated the genome editing efficiency in Bacteroides thetaiotaomicron, including the mode of Cas protein expression (constitutive, inducible), different Cas proteins (FnCas12a, SpRY, SpCas9) and sgRNAs. Researchers found that when Cas expression was controlled by the inducible promoter, FnCas12a displayed the highest efficiency of genome editing in B. thetaiotaomicron. This CRISPR/Cas-based tool could successfully delete large genomic fragments (up to 50 kb) of metabolic gene clusters in B. thetaiotaomicron and also could achieve targeted and markerless insertions (Figure 2).
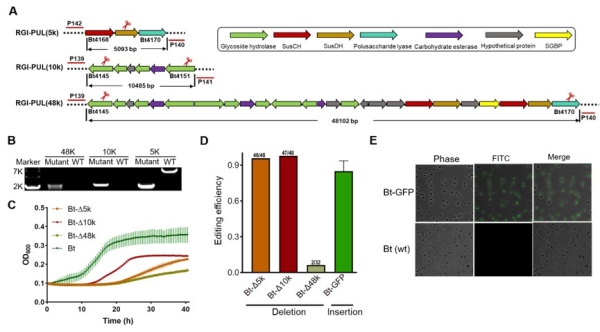
Fig. 2 Targeted deletions and insertions in B. thetaiotaomicron (Image by SIAT)
Finally, the new CRISPR/Cas-based genome editing tool was applied to edit the genomes of multiple Bacteroides species, including B.fragilis, B.ovatus, B.uniform and B.vulgatus (Figure 3). In addition, it is possible to obtain a markerless mutant by passaging in antibiotics-free medium for plasmid curing.
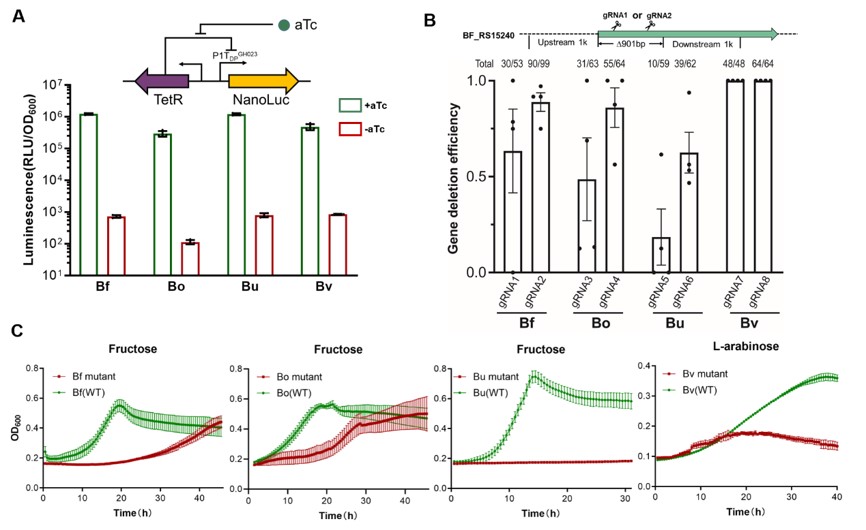
Fig. 3 Gene deletion in Bacteroides species (Image by SIAT)
In summary, the researchers developed CRISPR/Cas-based systems for versatile and highly efficient genetic manipulation of Bacteroides species, allowing targeted, markerless deletions and insertions in the genome. By laying a technical foundation for further understanding the function of human gut commensal bacteria and the development of engineered live bacteria therapeutics, this novel genome editing method for and will greatly facilitate basic and applied research on gut microbiota and human health.
Media Contact:
ZHANG Xiaomin
Email:xm.zhang@siat.ac.cn